Gated Graph Sequence Neural Networks
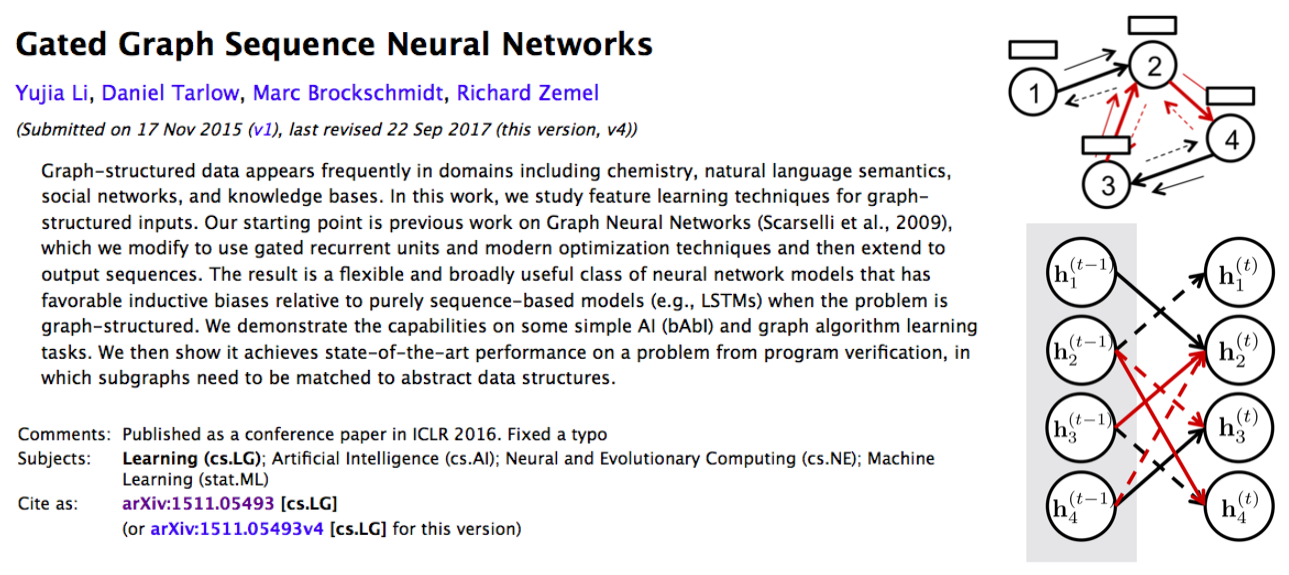
Graph-structured data appears frequently in domains including chemistry, natural language semantics, social networks, and knowledge bases. In this work, we study feature learning techniques for graph-structured inputs. Our starting point is previous work on Graph Neural Networks (Scarselli et al., 2009), which we modify to use gated recurrent units and modern optimization techniques and then extend to output sequences. The result is a flexible and broadly useful class of neural network models that has favorable inductive biases relative to purely sequence-based models (e.g., LSTMs) when the problem is graph-structured. We demonstrate the capabilities on some simple AI (bAbI) and graph algorithm learning tasks. We then show it achieves state-of-the-art performance on a problem from program verification, in which subgraphs need to be matched to abstract data structures.
PDF AbstractCode
| Task | Dataset | Model | Metric Name | Metric Value | Global Rank | Benchmark |
|---|---|---|---|---|---|---|
| Node Classification | CiteSeer (1%) | GGNN | Accuracy | 56.0% | # 11 | |
| Node Classification | CiteSeer with Public Split: fixed 20 nodes per class | GGNN | Accuracy | 64.6% | # 37 | |
| Node Classification | Cora (0.5%) | GGNN | Accuracy | 48.2% | # 12 | |
| Node Classification | Cora (1%) | GGNN | Accuracy | 60.5% | # 11 | |
| Node Classification | Cora (3%) | GGNN | Accuracy | 73.1% | # 11 | |
| Node Classification | Cora with Public Split: fixed 20 nodes per class | GGNN | Accuracy | 77.6% | # 32 | |
| Graph Classification | IPC-grounded | GG-NN | Accuracy | 77.9% | # 1 | |
| Graph Classification | IPC-lifted | GG-NN | Accuracy | 81.4% | # 2 | |
| Node Classification | PubMed (0.03%) | GGNN | Accuracy | 55.8% | # 11 | |
| Node Classification | PubMed (0.05%) | GGNN | Accuracy | 63.3% | # 10 | |
| Node Classification | PubMed (0.1%) | GGNN | Accuracy | 70.4% | # 10 | |
| Node Classification | PubMed with Public Split: fixed 20 nodes per class | GGNN | Accuracy | 75.8% | # 31 | |
| Drug Discovery | QM9 | Gated Graph Sequence NN | Error ratio | 1.36 | # 10 | |
| SQL-to-Text | WikiSQL | GGS-NN | BLEU-4 | 35.53 | # 2 |




















 Cora
Cora
 WikiSQL
WikiSQL
 bAbI
bAbI
 QM9
QM9
