G-CASCADE: Efficient Cascaded Graph Convolutional Decoding for 2D Medical Image Segmentation
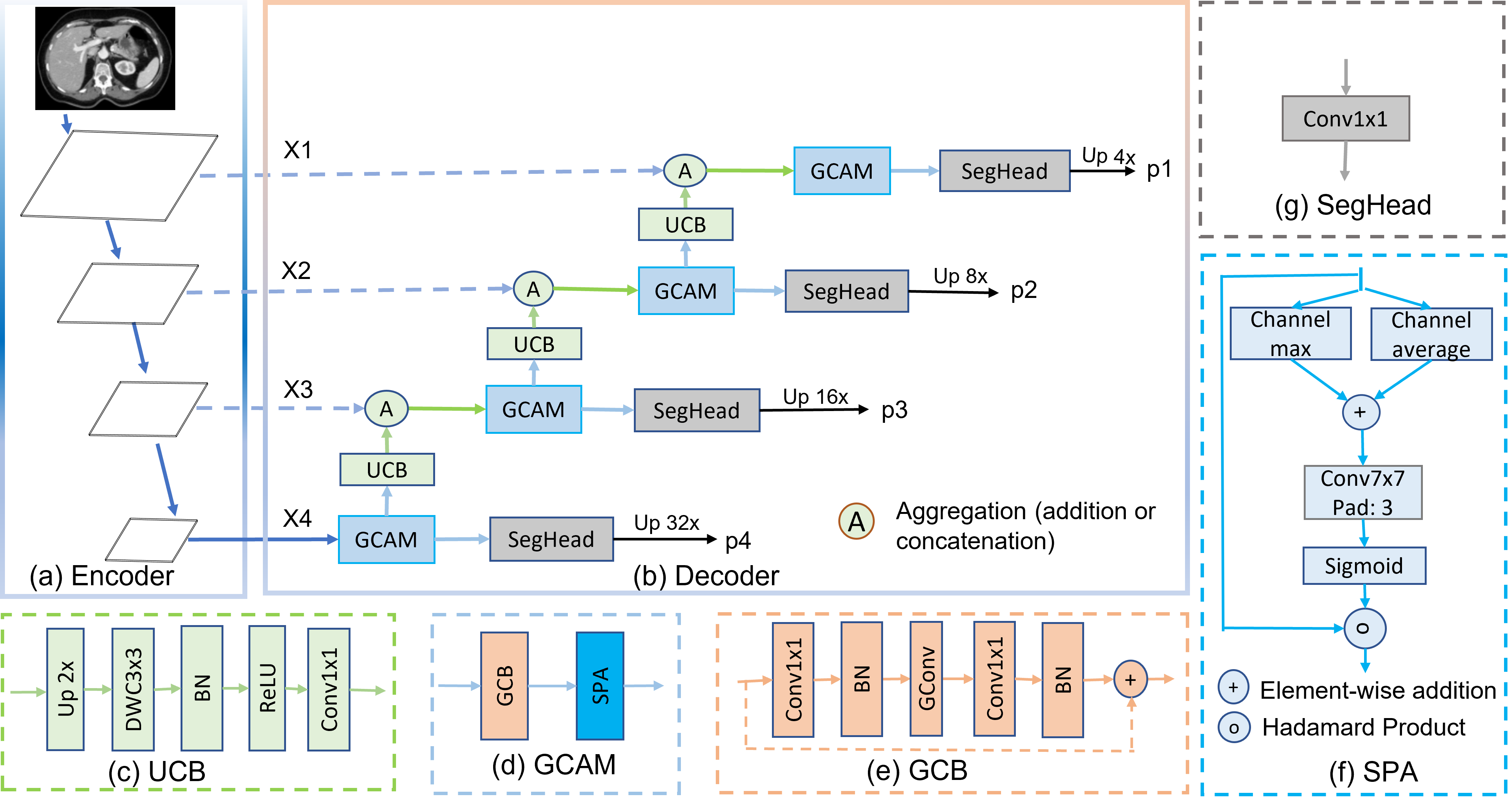
In recent years, medical image segmentation has become an important application in the field of computer-aided diagnosis. In this paper, we are the first to propose a new graph convolution-based decoder namely, Cascaded Graph Convolutional Attention Decoder (G-CASCADE), for 2D medical image segmentation. G-CASCADE progressively refines multi-stage feature maps generated by hierarchical transformer encoders with an efficient graph convolution block. The encoder utilizes the self-attention mechanism to capture long-range dependencies, while the decoder refines the feature maps preserving long-range information due to the global receptive fields of the graph convolution block. Rigorous evaluations of our decoder with multiple transformer encoders on five medical image segmentation tasks (i.e., Abdomen organs, Cardiac organs, Polyp lesions, Skin lesions, and Retinal vessels) show that our model outperforms other state-of-the-art (SOTA) methods. We also demonstrate that our decoder achieves better DICE scores than the SOTA CASCADE decoder with 80.8% fewer parameters and 82.3% fewer FLOPs. Our decoder can easily be used with other hierarchical encoders for general-purpose semantic and medical image segmentation tasks.
PDF AbstractCode
Results from the Paper
 Ranked #1 on
Retinal Vessel Segmentation
on DRIVE
(Specificity metric)
Ranked #1 on
Retinal Vessel Segmentation
on DRIVE
(Specificity metric)















 DRIVE
DRIVE
 Kvasir-SEG
Kvasir-SEG
 Kvasir
Kvasir
 CHASE_DB1
CHASE_DB1
 ACDC
ACDC
 CVC-ClinicDB
CVC-ClinicDB
 ISIC 2018 Task 1
ISIC 2018 Task 1
 MICCAI 2015 Multi-Atlas Abdomen Labeling Challenge
MICCAI 2015 Multi-Atlas Abdomen Labeling Challenge