Dual-Query Multiple Instance Learning for Dynamic Meta-Embedding based Tumor Classification
Whole slide image (WSI) assessment is a challenging and crucial step in cancer diagnosis and treatment planning. WSIs require high magnifications to facilitate sub-cellular analysis. Precise annotations for patch- or even pixel-level classifications in the context of gigapixel WSIs are tedious to acquire and require domain experts. Coarse-grained labels, on the other hand, are easily accessible, which makes WSI classification an ideal use case for multiple instance learning (MIL). In our work, we propose a novel embedding-based Dual-Query MIL pipeline (DQ-MIL). We contribute to both the embedding and aggregation steps. Since all-purpose visual feature representations are not yet available, embedding models are currently limited in terms of generalizability. With our work, we explore the potential of dynamic meta-embedding based on cutting-edge self-supervised pre-trained models in the context of MIL. Moreover, we propose a new MIL architecture capable of combining MIL-attention with correlated self-attention. The Dual-Query Perceiver design of our approach allows us to leverage the concept of self-distillation and to combine the advantages of a small model in the context of a low data regime with the rich feature representation of a larger model. We demonstrate the superior performance of our approach on three histopathological datasets, where we show improvement of up to 10% over state-of-the-art approaches.
PDF Abstract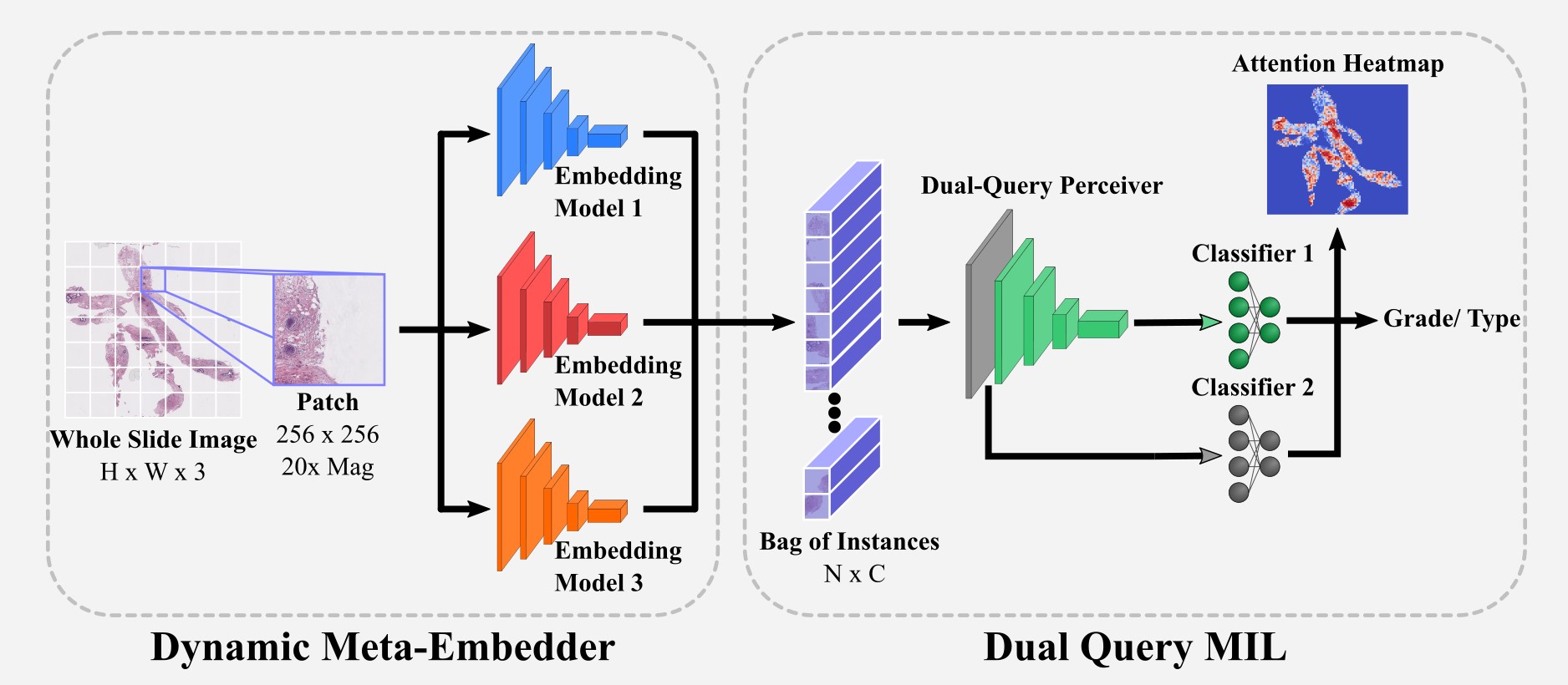


 ImageNet
ImageNet