Deep Inverse Reinforcement Learning for Structural Evolution of Small Molecules
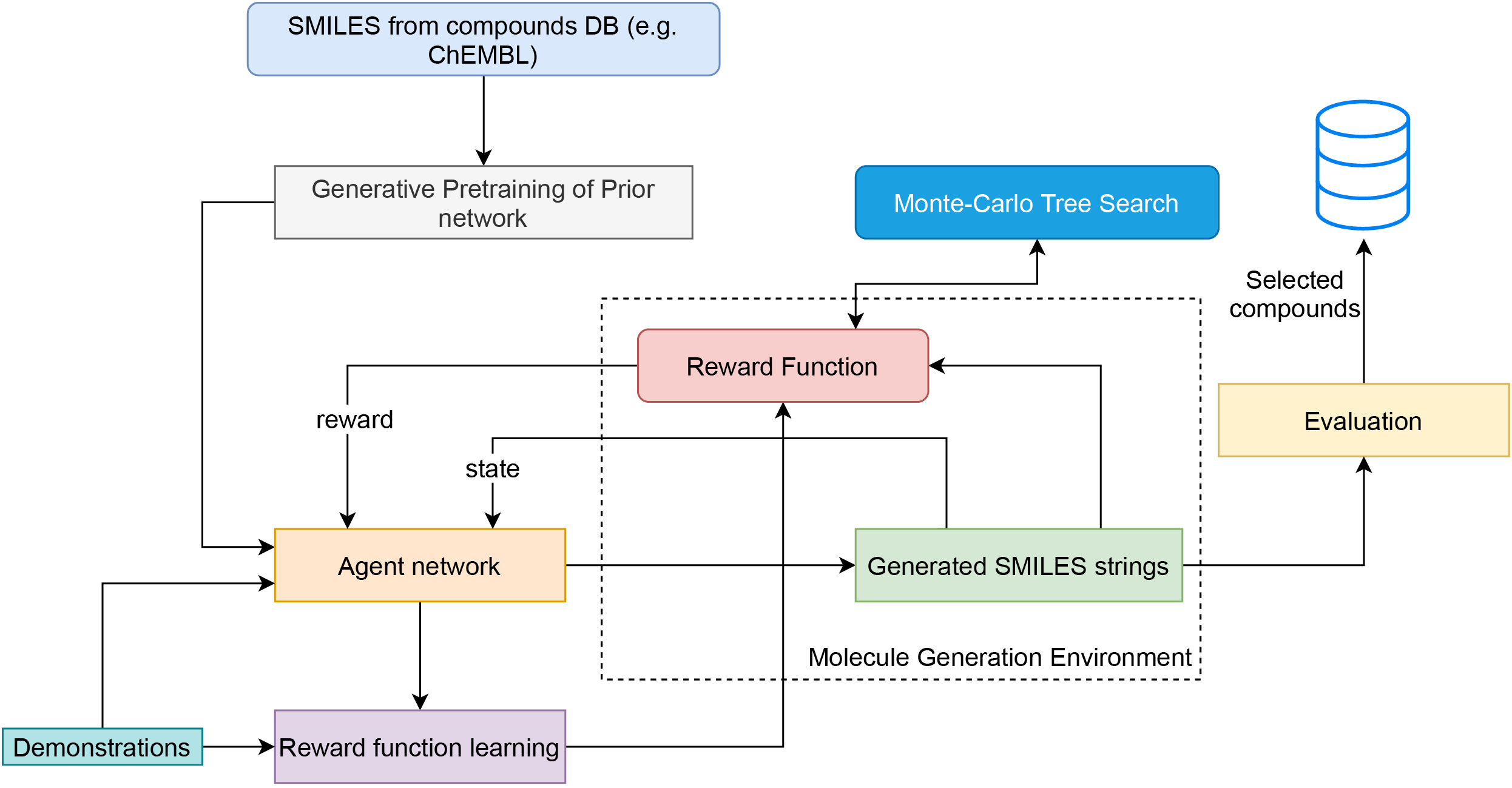
The size and quality of chemical libraries to the drug discovery pipeline are crucial for developing new drugs or repurposing existing drugs. Existing techniques such as combinatorial organic synthesis and High-Throughput Screening usually make the process extraordinarily tough and complicated since the search space of synthetically feasible drugs is exorbitantly huge. While reinforcement learning has been mostly exploited in the literature for generating novel compounds, the requirement of designing a reward function that succinctly represents the learning objective could prove daunting in certain complex domains. Generative Adversarial Network-based methods also mostly discard the discriminator after training and could be hard to train. In this study, we propose a framework for training a compound generator and learning a transferable reward function based on the entropy maximization inverse reinforcement learning paradigm. We show from our experiments that the inverse reinforcement learning route offers a rational alternative for generating chemical compounds in domains where reward function engineering may be less appealing or impossible while data exhibiting the desired objective is readily available.
PDF Abstract