Semi-Supervised Classification with Graph Convolutional Networks
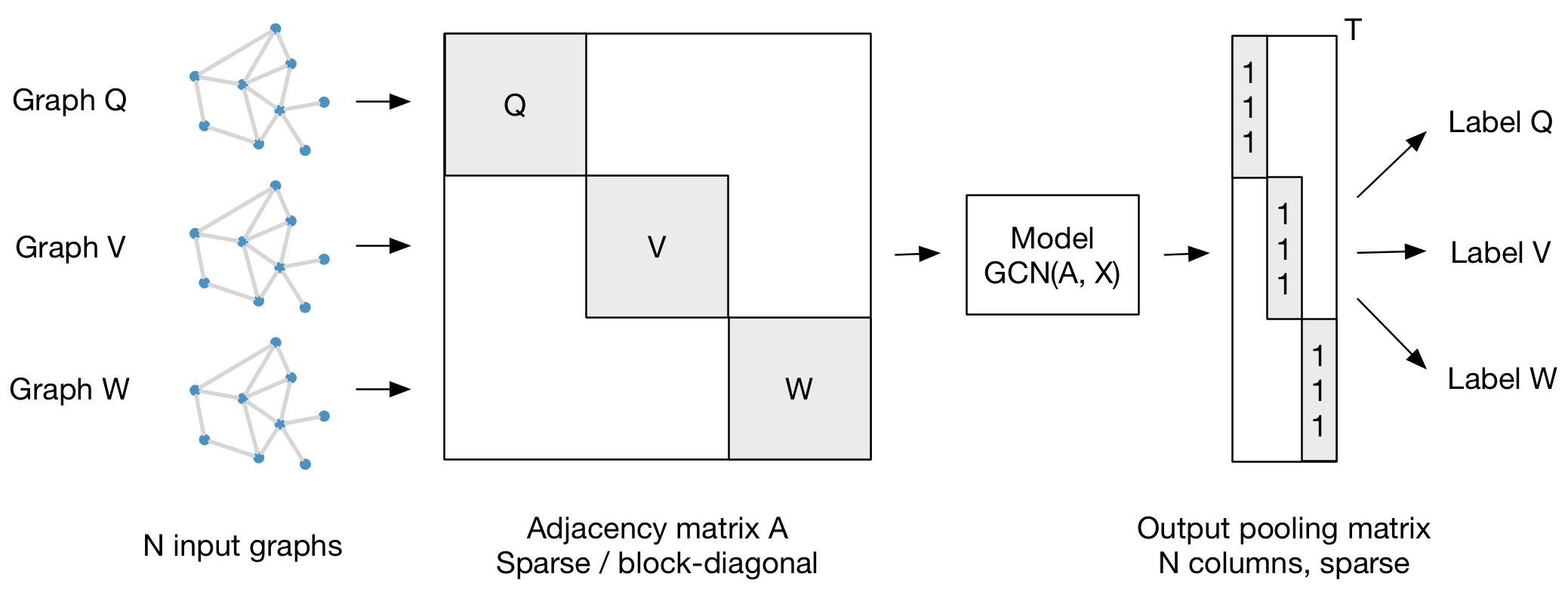
We present a scalable approach for semi-supervised learning on graph-structured data that is based on an efficient variant of convolutional neural networks which operate directly on graphs. We motivate the choice of our convolutional architecture via a localized first-order approximation of spectral graph convolutions. Our model scales linearly in the number of graph edges and learns hidden layer representations that encode both local graph structure and features of nodes. In a number of experiments on citation networks and on a knowledge graph dataset we demonstrate that our approach outperforms related methods by a significant margin.
PDF AbstractCode
Datasets
 OGB
OGB
 Flickr30k
Flickr30k
 Cora
Citeseer
Cora
Citeseer
 NELL
Penn94
genius
twitch-gamers
Deezer-Europe
Wisconsin(60%/20%/20% random splits)
Squirrel (60%/20%/20% random splits)
Texas(60%/20%/20% random splits)
Film (60%/20%/20% random splits)
Cornell (60%/20%/20% random splits)
PubMed (60%/20%/20% random splits)
Chameleon(60%/20%/20% random splits)
NELL
Penn94
genius
twitch-gamers
Deezer-Europe
Wisconsin(60%/20%/20% random splits)
Squirrel (60%/20%/20% random splits)
Texas(60%/20%/20% random splits)
Film (60%/20%/20% random splits)
Cornell (60%/20%/20% random splits)
PubMed (60%/20%/20% random splits)
Chameleon(60%/20%/20% random splits)
 PCQM4Mv2-LSC
Brazil Air-Traffic
PCQM4Mv2-LSC
Brazil Air-Traffic
 Drug Combination Extraction Dataset
Drug Combination Extraction Dataset